2F8B
 
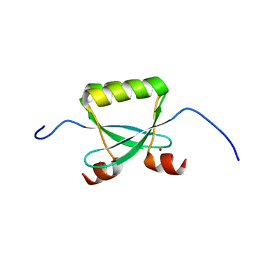 | |
2EWL
 
 | |
1HO2
 
 | | NMR STRUCTURE OF THE POTASSIUM CHANNEL FRAGMENT L45 IN MICELLES | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL PROTEIN | | Authors: | Ohlenschlager, O, Hojo, H, Ramachandran, R, Gorlach, M, Haris, P.I. | | Deposit date: | 2000-12-08 | | Release date: | 2002-06-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the S4-S5 segment of the Shaker potassium channel.
Biophys.J., 82, 2002
|
|
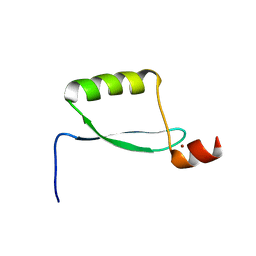
1HO7
 
 | | NMR STRUCTURE OF THE POTASSIUM CHANNEL FRAGMENT L45 IN TFE | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL PROTEIN | | Authors: | Ohlenschlager, O, Hojo, H, Ramachandran, R, Gorlach, M, Haris, P.I. | | Deposit date: | 2000-12-10 | | Release date: | 2002-06-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the S4-S5 segment of the Shaker potassium channel.
Biophys.J., 82, 2002
|
|
1RFR
 
 | | NMR structure of the 30mer stemloop-D of coxsackieviral RNA | | Descriptor: | stemloop-D RNA of the 5'-cloverleaf of coxsackievirus B3 | | Authors: | Ohlenschlager, O, Wohnert, J, Bucci, E, Seitz, S, Hafner, S, Ramachandran, R, Zell, R, Gorlach, M. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-23 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf
RNA and its interaction with the proteinase 3C.
STRUCTURE, 12, 2004
|
|
1SSN
 
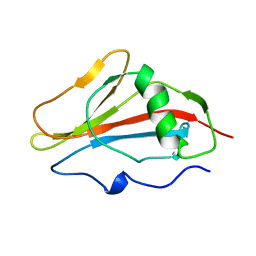 | | STAPHYLOKINASE, SAKSTAR VARIANT, NMR, 20 STRUCTURES | | Descriptor: | STAPHYLOKINASE | | Authors: | Ohlenschlager, O, Ramachandran, R, Guhrs, K.H, Schlott, B, Brown, L.R. | | Deposit date: | 1998-06-07 | | Release date: | 1998-12-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the plasminogen-activator protein staphylokinase.
Biochemistry, 37, 1998
|
|
2KMU
 
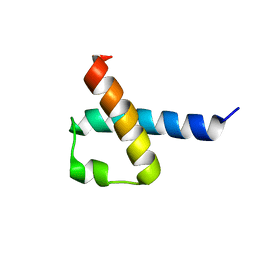 | |
2JYM
 
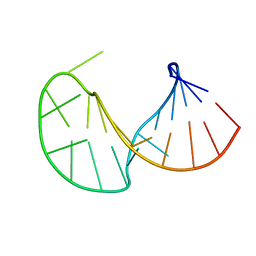 | |
1D6K
 
 | | NMR SOLUTION STRUCTURE OF THE 5S RRNA E-LOOP/L25 COMPLEX | | Descriptor: | 5S RRNA E-LOOP (5SE), RIBOSOMAL PROTEIN L25 | | Authors: | Stoldt, M, Wohnert, J, Ohlenschlager, O, Gorlach, M, Brown, L.R. | | Deposit date: | 1999-10-14 | | Release date: | 1999-11-22 | | Last modified: | 2022-03-23 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the 5S rRNA E-domain-protein L25 complex shows preformed and induced recognition.
EMBO J., 18, 1999
|
|
1Z30
 
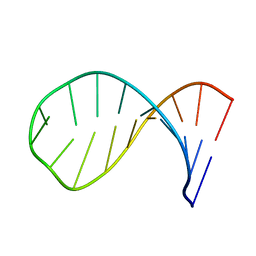 | | NMR structure of the apical part of stemloop D from cloverleaf 1 of bovine enterovirus 1 RNA | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*UP*CP*GP*UP*UP*AP*GP*AP*AP*CP*GP*UP*C)-3' | | Authors: | Ihle, Y, Ohlenschlager, O, Duchardt, E, Ramachandran, R, Gorlach, M. | | Deposit date: | 2005-03-10 | | Release date: | 2005-04-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A novel cGUUAg tetraloop structure with a conserved yYNMGg-type backbone conformation from cloverleaf 1 of bovine enterovirus 1 RNA
Nucleic Acids Res., 33, 2005
|
|
1UUU
 
 | | STRUCTURE OF AN RNA HAIRPIN LOOP WITH A 5'-CGUUUCG-3' LOOP MOTIF BY HETERONUCLEAR NMR SPECTROSCOPY AND DISTANCE GEOMETRY, 15 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*AP*CP*GP*UP*UP*UP*CP*GP*UP*AP*CP*GP*CP*C)-3') | | Authors: | Sich, C, Ohlenschlager, O, Ramachandran, R, Gorlach, M, Brown, L.R. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin loop with a 5'-CGUUUCG-3' loop motif by heteronuclear NMR spectroscopy and distance geometry.
Biochemistry, 36, 1997
|
|
1KMA
 
 | | NMR Structure of the Domain-I of the Kazal-type Thrombin Inhibitor Dipetalin | | Descriptor: | DIPETALIN | | Authors: | Schlott, B, Wohnert, J, Icke, C, Hartmann, M, Ramachandran, R, Guhrs, K.-H, Glusa, E, Flemming, J, Gorlach, M, Grosse, F, Ohlenschlager, O. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Interaction of Kazal-type inhibitor domains with serine proteinases: biochemical and structural studies.
J.Mol.Biol., 318, 2002
|
|
6GZR
 
 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | Descriptor: | 5-carboxy methylrhodamine, tetramethylrhodamine aptamer | | Authors: | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-17 | | Last modified: | 2020-01-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
6GZK
 
 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | Descriptor: | 5-carboxy methylrhodamine, TMR3 (48-MER) | | Authors: | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | Deposit date: | 2018-07-04 | | Release date: | 2019-07-17 | | Last modified: | 2020-01-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
1EE7
 
 | | NMR STRUCTURE OF THE PEPTAIBOL CHRYSOSPERMIN C BOUND TO DPC MICELLES | | Descriptor: | CHRYSOSPERMIN C | | Authors: | Anders, R, Ohlenschlager, O, Soskic, V, Wenschuh, H, Heise, B, Brown, L.R. | | Deposit date: | 2000-01-31 | | Release date: | 2000-05-10 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The NMR Solution Structure of the Ion Channel Peptaibol Chrysospermin C Bound to Dodecylphosphocholine Micelles.
Eur.J.Biochem., 267, 2000
|
|
1OKD
 
 | | NMR-structure of tryparedoxin 1 | | Descriptor: | TRYPAREDOXIN 1 | | Authors: | Krumme, D, Budde, H, Hecht, H.-J, Menge, U, Ohlenschlager, O, Ross, A, Wissing, J, Wray, V, Flohe, L. | | Deposit date: | 2003-07-22 | | Release date: | 2003-08-28 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | NMR studies of the interaction of tryparedoxin with redox-inactive substrate homologues.
Biochemistry, 42, 2003
|
|
1RQM
 
 | | SOLUTION STRUCTURE OF THE K18G/R82E ALICYCLOBACILLUS ACIDOCALDARIUS THIOREDOXIN MUTANT | | Descriptor: | Thioredoxin | | Authors: | Leone, M, Di Lello, P, Ohlenschlager, O, Pedone, E.M, Bartolucci, S, Rossi, M, Di Blasio, B, Pedone, C, Saviano, M, Isernia, C, Fattorusso, R. | | Deposit date: | 2003-12-05 | | Release date: | 2004-06-22 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the K18G/R82E Alicyclobacillus acidocaldarius Thioredoxin Mutant: A Molecular Analysis of Its Reduced Thermal Stability.
Biochemistry, 43, 2004
|
|
1S9L
 
 | | NMR Solution Structure of a Parallel LNA Quadruplex | | Descriptor: | 5'-((TLN)P*(LCG)P*(LCG)P*(LCG)P*(TLN))-3' | | Authors: | Randazzo, A, Esposito, V, Ohlenschlager, O, Ramachandran, R, Mayola, L. | | Deposit date: | 2004-02-05 | | Release date: | 2004-06-29 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a parallel LNA quadruplex.
Nucleic Acids Res., 32, 2004
|
|
2K8D
 
 | |
2KXM
 
 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostmycin complex | | Descriptor: | RIBOSTAMYCIN, RNA (27-MER) | | Authors: | Duchardt-Ferner, E, Weigand, J.E, Ohlenschlager, O, Schmidtke, S.R, Suess, B, Wohnert, J. | | Deposit date: | 2010-05-10 | | Release date: | 2011-04-20 | | Last modified: | 2014-02-05 | | Method: | SOLUTION NMR | | Cite: | Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2M3M
 
 | |
2M3L
 
 | |
2M45
 
 | | NMR solution structure of the C-terminus of the minichromosome maintenance protein MCM from Sulfolobus solfataricus | | Descriptor: | Minichromosome maintenance protein MCM | | Authors: | Wiedemann, C, Ohlenschlager, O, Medagli, B, Onesti, S, Gorlach, M. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and regulatory role of the C-terminal winged helix domain of the archaeal minichromosome maintenance complex
Nucleic Acids Res., 43, 2015
|
|
2MA3
 
 | | NMR solution structure of the C-terminus of the minichromosome maintenance protein MCM from Methanothermobacter thermautotrophicus | | Descriptor: | DNA replication initiator (Cdc21/Cdc54) | | Authors: | Wiedemann, C, Ohlenschlager, O, Medagli, B, Onesti, S, Gorlach, M. | | Deposit date: | 2013-06-26 | | Release date: | 2014-12-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and regulatory role of the C-terminal winged helix domain of the archaeal minichromosome maintenance complex.
Nucleic Acids Res., 43, 2015
|
|
5LWJ
 
 | | Solution NMR structure of the GTP binding Class II RNA aptamer-ligand-complex containing a protonated adenine nucleotide with a highly shifted pKa. | | Descriptor: | GTP Class II RNA (34-MER), GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wolter, A.C, Weickhmann, A.K, Nasiri, A.H, Hantke, K, Ohlenschlaeger, O, Wunderlich, C.H, Kreutz, C, Duchardt-Ferner, E, Woehnert, J. | | Deposit date: | 2016-09-17 | | Release date: | 2016-12-14 | | Last modified: | 2019-09-18 | | Method: | SOLUTION NMR | | Cite: | A Stably Protonated Adenine Nucleotide with a Highly Shifted pKa Value Stabilizes the Tertiary Structure of a GTP-Binding RNA Aptamer.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
