8QN3
 
 | | OPR3 wildtype in complex with NADH4 | | Descriptor: | 1,4,5,6-Tetrahydronicotinamide adenine dinucleotide, 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Bijelic, A, Macheroux, P, Keschbaumer, B. | | Deposit date: | 2023-09-25 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Loop 6 and the beta-hairpin flap are structural hotspots that determine cofactor specificity in the FMN-dependent family of ene-reductases.
Febs J., 291, 2024
|
|
8QMX
 
 | | OPR3 wildtype in complex with NADPH4 | | Descriptor: | 12-oxophytodienoate reductase 3, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2023-09-25 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Loop 6 and the beta-hairpin flap are structural hotspots that determine cofactor specificity in the FMN-dependent family of ene-reductases.
Febs J., 291, 2024
|
|
5A4K
 
 | | Crystal structure of the R139W variant of human NAD(P)H:quinone oxidoreductase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Strandback, E, Gudipati, V, Uhl, M.K, Rantase, D.M, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2015-06-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Catalytic competence, structure and stability of the cancer-associated R139W variant of the human NAD(P)H:quinone oxidoreductase 1 (NQO1).
FEBS J., 284, 2017
|
|
2YHE
 
 | | Structure determination of the stereoselective inverting sec- alkylsulfatase Pisa1 from Pseudomonas sp. | | Descriptor: | SEC-ALKYL SULFATASE, SULFATE ION, ZINC ION | | Authors: | Kepplinger, B, Faber, K, Macheroux, P, Schober, M, Knaus, T, Wagner, U.G. | | Deposit date: | 2011-04-29 | | Release date: | 2012-05-16 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Mechanism of an Inverting Alkylsulfatase from Pseudomonas Sp. Dsm6611 Specific for Secondary Alkylsulfates.
FEBS J., 279, 2012
|
|
1F8R
 
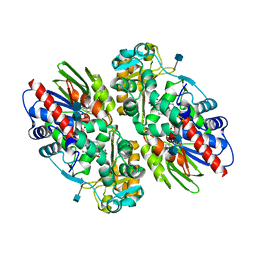 | | CRYSTAL STRUCTURE OF L-AMINO ACID OXIDASE FROM CALLOSELASMA RHODOSTOMA COMPLEXED WITH CITRATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Pawelek, P.D, Cheah, J, Coulombe, R, Macheroux, P, Ghisla, S, Vrielink, A. | | Deposit date: | 2000-07-04 | | Release date: | 2000-08-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site.
EMBO J., 19, 2000
|
|
1F8S
 
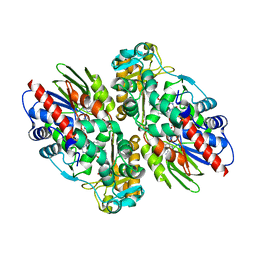 | | CRYSTAL STRUCTURE OF L-AMINO ACID OXIDASE FROM CALLOSELASMA RHODOSTOMA, COMPLEXED WITH THREE MOLECULES OF O-AMINOBENZOATE. | | Descriptor: | 2-AMINOBENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pawelek, P.D, Cheah, J, Coulombe, R, Macheroux, P, Ghisla, S, Vrielink, A. | | Deposit date: | 2000-07-04 | | Release date: | 2000-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site.
EMBO J., 19, 2000
|
|
4PWC
 
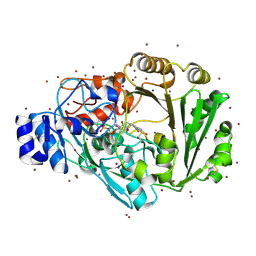 | | Phl p 4 I153V N158H variant, a glucose oxidase, 3.5 M NaBr soak | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Pollen allergen Phl p 4.0202, ... | | Authors: | Zafred, D, Keller, W, Macheroux, P. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rationally engineered flavin-dependent oxidase reveals steric control of dioxygen reduction.
Febs J., 282, 2015
|
|
4PVK
 
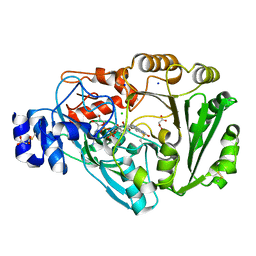 | | Phl p 4 I153V N158H variant, a glucose oxidase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, ... | | Authors: | Zafred, D, Teufelberger, A, Keller, W, Macheroux, P. | | Deposit date: | 2014-03-17 | | Release date: | 2014-04-02 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Rationally engineered flavin-dependent oxidase reveals steric control of dioxygen reduction.
Febs J., 282, 2015
|
|
5E33
 
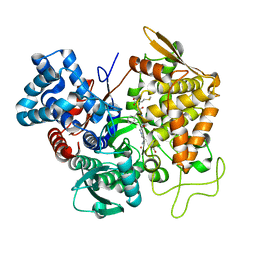 | | Structure of human DPP3 in complex with met-enkephalin | | Descriptor: | Dipeptidyl peptidase 3, MAGNESIUM ION, Met-enkephalin, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-01 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.837 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5E3A
 
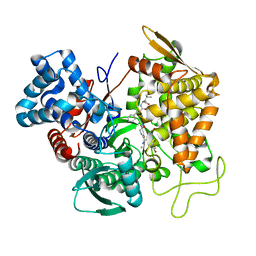 | | Structure of human DPP3 in complex with opioid peptide leu-enkephalin | | Descriptor: | Dipeptidyl peptidase 3, Leu-enkephalin, MAGNESIUM ION, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5E3C
 
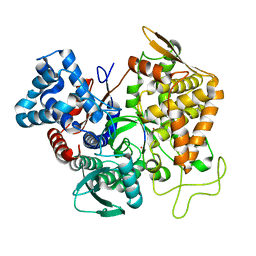 | | Structure of human DPP3 in complex with hemorphin like opioid peptide IVYPW | | Descriptor: | Dipeptidyl peptidase 3, IVYPW, MAGNESIUM ION, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.765 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5EGY
 
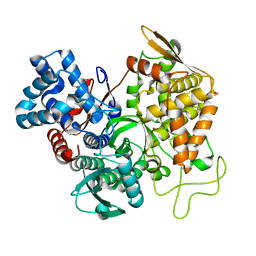 | | Structure of ligand free human DPP3 in closed form. | | Descriptor: | Dipeptidyl peptidase 3, MAGNESIUM ION, ZINC ION | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.741 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5EHH
 
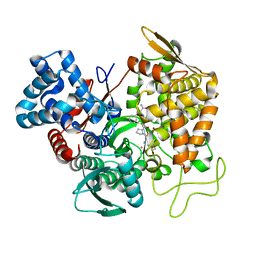 | | Structure of human DPP3 in complex with endomorphin-2. | | Descriptor: | Dipeptidyl peptidase 3, Endomorphin-2, MAGNESIUM ION, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-28 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
1ICQ
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH 9R,13R-OPDA | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, 9R,13R-12-OXOPHYTODIENOIC ACID, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1ICP
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH PEG400 | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1ICS
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
3D2J
 
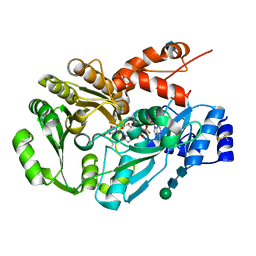 | | Structure of berberine bridge enzyme from Eschscholzia californica, tetragonal crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Winkler, A, Lyskowski, A, Macheroux, P, Gruber, K. | | Deposit date: | 2008-05-08 | | Release date: | 2008-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A concerted mechanism for berberine bridge enzyme
Nat.Chem.Biol., 4, 2008
|
|
3D2H
 
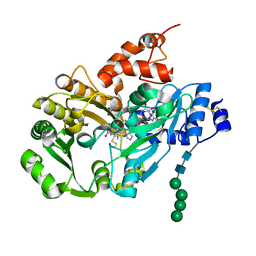 | | Structure of berberine bridge enzyme from Eschscholzia californica, monoclinic crystal form | | Descriptor: | (2R,3S,4S)-5-[(4R)-6',7'-dimethyl-2,3',5-trioxo-1'H-spiro[imidazolidine-4,2'-quinoxalin]-4'(3'H)-yl]-2,3,4-trihydroxypentyl-adenosine diphosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winkler, A, Lyskowski, A, Macheroux, P, Gruber, K. | | Deposit date: | 2008-05-08 | | Release date: | 2008-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A concerted mechanism for berberine bridge enzyme
Nat.Chem.Biol., 4, 2008
|
|
3CSK
 
 | | Structure of DPP III from Saccharomyces cerevisiae | | Descriptor: | MAGNESIUM ION, Probable dipeptidyl-peptidase 3, ZINC ION | | Authors: | Baral, P.K, Jajcanin, N, Deller, S, Macheroux, P, Abramic, M, Gruber, K. | | Deposit date: | 2008-04-10 | | Release date: | 2008-06-10 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The first structure of dipeptidyl-peptidase III provides insight into the catalytic mechanism and mode of substrate binding.
J.Biol.Chem., 283, 2008
|
|
3D2D
 
 | | Structure of berberine bridge enzyme in complex with (S)-reticuline | | Descriptor: | (S)-reticuline, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winkler, A, Lyskowski, A, Macheroux, P, Gruber, K. | | Deposit date: | 2008-05-08 | | Release date: | 2008-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | A concerted mechanism for berberine bridge enzyme
Nat.Chem.Biol., 4, 2008
|
|
4PVH
 
 | | Phl p 4 N158H variant, a glucose dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, Pollen allergen Phl p 4.0202, ... | | Authors: | Zafred, D, Teufelberger, A, Keller, W, Macheroux, P. | | Deposit date: | 2014-03-17 | | Release date: | 2014-04-02 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rationally engineered flavin-dependent oxidase reveals steric control of dioxygen reduction.
Febs J., 282, 2015
|
|
4PWB
 
 | | Phl p 4 I153V variant, a glucose oxidase, pressurized with Xenon | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, Pollen allergen Phl p 4.0202, ... | | Authors: | Zafred, D, Keller, W, Macheroux, P. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rationally engineered flavin-dependent oxidase reveals steric control of dioxygen reduction.
Febs J., 282, 2015
|
|
4PZF
 
 | | Berberine bridge enzyme G164A variant, a reticuline dehydrogenase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DODECAETHYLENE GLYCOL, ... | | Authors: | Zafred, D, Wallner, S, Steiner, B, Macheroux, P. | | Deposit date: | 2014-03-30 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rationally engineered flavin-dependent oxidase reveals steric control of dioxygen reduction.
Febs J., 282, 2015
|
|
4PVJ
 
 | | Phl p 4 I153V variant, a glucose oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, Pollen allergen Phl p 4.0202, ... | | Authors: | Zafred, D, Teufelberger, A, Keller, W, Macheroux, P. | | Deposit date: | 2014-03-17 | | Release date: | 2014-04-02 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rationally engineered flavin-dependent oxidase reveals steric control of dioxygen reduction.
Febs J., 282, 2015
|
|
4PVE
 
 | | Wild-type Phl p 4.0202, a glucose dehydrogenase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, ... | | Authors: | Zafred, D, Teufelberger, A, Keller, W, Macheroux, P. | | Deposit date: | 2014-03-17 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rationally engineered flavin-dependent oxidase reveals steric control of dioxygen reduction.
Febs J., 282, 2015
|
|
