2MWK
 
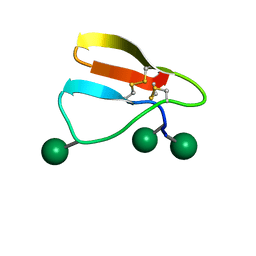 | | Family 1 Carbohydrate-Binding Module from Trichoderma reesei Cel7A with O-mannose residues at Thr1, Ser3, and Ser14 | | Descriptor: | Exoglucanase 1, alpha-D-mannopyranose | | Authors: | Happs, R.M, Chen, L, Resch, M.G, Davis, M.F, Beckham, G.T, Tan, Z, Crowley, M.F. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-02 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | O-glycosylation effects on family 1 carbohydrate-binding module solution structures.
Febs J., 282, 2015
|
|
2MWJ
 
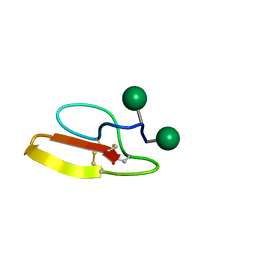 | | Solution structure of Family 1 Carbohydrate-Binding Module from Trichoderma reesei Cel7A with O-mannose residues at Thr1 and Ser3 | | Descriptor: | Exoglucanase 1, alpha-D-mannopyranose | | Authors: | Happs, R.M, Chen, L, Resch, M.G, Davis, M.F, Beckham, G.T, Tan, Z, Crowley, M.F. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-02 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | O-glycosylation effects on family 1 carbohydrate-binding module solution structures.
Febs J., 282, 2015
|
|
4B5Q
 
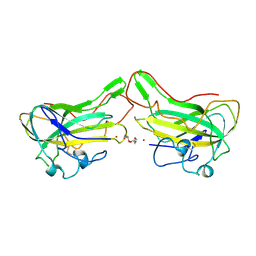 | | The lytic polysaccharide monooxygenase GH61D structure from the basidiomycota fungus Phanerochaete chrysosporium | | Descriptor: | COPPER (II) ION, GLYCEROL, GLYCOSIDE HYDROLASE FAMILY 61 PROTEIN D, ... | | Authors: | Wu, M, Beckham, G.T, Larsson, A.M, Ishida, T, Kim, S, Crowley, M.F, Payne, C.M, Horn, S.J, Westereng, B, Stahlberg, J, Eijsink, V.G.H, Sandgren, M. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Computational Characterization of the Lytic Polysaccharide Monooxygenase Gh61D from the Basidiomycota Fungus Phanerochaete Chrysosporium
J.Biol.Chem., 288, 2013
|
|
4XEB
 
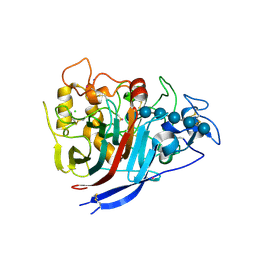 | | The structure of P. funicolosum Cel7A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glucanase, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2014-12-23 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering enhanced cellobiohydrolase activity
Nat Commun, 9(1), 2018
|
|
5OMS
 
 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with guaethol. | | Descriptor: | 2-ethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5OMR
 
 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with vanillin. | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, GcoA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5BV9
 
 | |
6HQQ
 
 | | Crystal structure of GcoA F169A bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQN
 
 | | Crystal structure of GcoA F169L bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQL
 
 | | Crystal structure of GcoA F169H bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQT
 
 | | Crystal structure of GcoA F169V bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQO
 
 | | Crystal structure of GcoA F169S bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5KX6
 
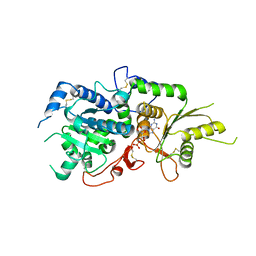 | | The structure of Arabidopsis thaliana FUT1 Mutant R284K in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water-mediated mechanism.
Plant J., 91, 2017
|
|
5KWK
 
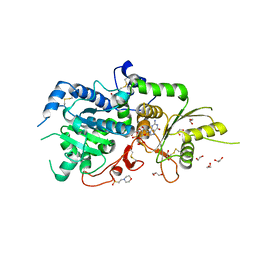 | | The structure of Arabidopsis thaliana FUT1 in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2016-07-18 | | Release date: | 2016-09-28 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water-mediated mechanism.
Plant J., 91, 2017
|
|
5KOE
 
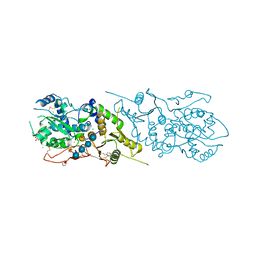 | | The structure of Arabidopsis thaliana FUT1 in complex with XXLG | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water-mediated mechanism.
Plant J., 91, 2017
|
|
5NCB
 
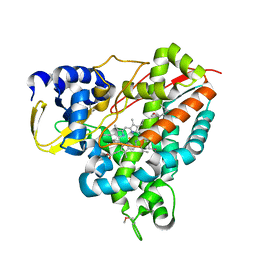 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with guaiacol. | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-07-04 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5NNS
 
 | | Crystal structure of HiLPMO9B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, COPPER (II) ION, ... | | Authors: | Dimarogona, M, Sandgren, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and molecular dynamics studies of a C1-oxidizing lytic polysaccharide monooxygenase from Heterobasidion irregulare reveal amino acids important for substrate recognition.
FEBS J., 285, 2018
|
|
5OGX
 
 | | Crystal structure of Amycolatopsis cytochrome P450 reductase GcoB. | | Descriptor: | BROMIDE ION, Cytochrome P450 reductase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-07-13 | | Release date: | 2018-07-04 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5OMU
 
 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
4AVO
 
 | | Thermobifida fusca cellobiohydrolase Cel6B catalytic mutant D274A cocrystallized with cellobiose | | Descriptor: | ACETATE ION, BETA-1,4-EXOCELLULASE, CALCIUM ION, ... | | Authors: | Wu, M, Vuong, T.V, Wilson, D.B, Sandgren, M, Stahlberg, J, Hansson, H. | | Deposit date: | 2012-05-28 | | Release date: | 2013-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Loop Motions Important to Product Expulsion in the Thermobifida Fusca Glycoside Hydrolase Family 6 Cellobiohydrolase from Structural and Computational Studies.
J.Biol.Chem., 288, 2013
|
|
4AVN
 
 | | Thermobifida fusca cellobiohydrolase Cel6B catalytic mutant D226A- S232A cocrystallized with cellobiose | | Descriptor: | CALCIUM ION, CELLOBIOHYDROLASE. GLYCOSYL HYDROLASE FAMILY 6, beta-D-glucopyranose, ... | | Authors: | Wu, M, Vuong, T.V, Wilson, D.B, Sandgren, M, Stahlberg, J, Hansson, H. | | Deposit date: | 2012-05-28 | | Release date: | 2013-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Loop Motions Important to Product Expulsion in the Thermobifida Fusca Glycoside Hydrolase Family 6 Cellobiohydrolase from Structural and Computational Studies.
J.Biol.Chem., 288, 2013
|
|
4C4C
 
 | | Michaelis complex of Hypocrea jecorina CEL7A E217Q mutant with cellononaose spanning the active site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, COBALT (II) ION, ... | | Authors: | Haddad-Momeni, M, Sandgren, M, Stahlberg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Mechanism of Cellulose Hydrolysis by a Two-Step, Retaining Cellobiohydrolase Elucidated by Structural and Transition Path Sampling Studies.
J.Am.Chem.Soc., 136, 2014
|
|
4C4D
 
 | | Covalent glycosyl-enzyme intermediate of Hypocrea jecorina Cel7a E217Q mutant trapped using DNP-2-deoxy-2-fluoro-cellotrioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, COBALT (II) ION, ... | | Authors: | Haddad-Momeni, M, Mackenzie, L, Sandgren, M, Withers, S.G, Stahlberg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The Mechanism of Cellulose Hydrolysis by a Two-Step, Retaining Cellobiohydrolase Elucidated by Structural and Transition Path Sampling Studies.
J.Am.Chem.Soc., 136, 2014
|
|
6RWF
 
 | |
4DOD
 
 | | The structure of Cbescii CelA GH9 module | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 1,4-beta-glucanase, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2012-02-09 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA.
Science, 342, 2013
|
|
