5WKP
 
 | |
5WLW
 
 | |
5WGB
 
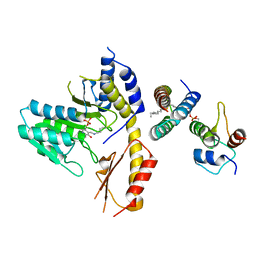 | |
7M8H
 
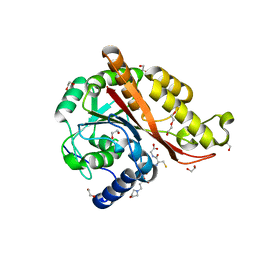 | |
7RTK
 
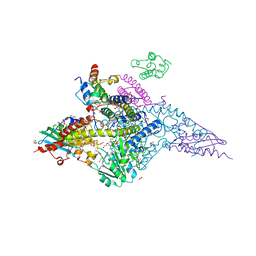 | | Structure of the (NIAU)2 complex with N-terminal mutation of ISCU2 Y35D at 2.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ... | | Authors: | Boniecki, M.T, Cygler, M. | | Deposit date: | 2021-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
To Be Published
|
|
7KQ8
 
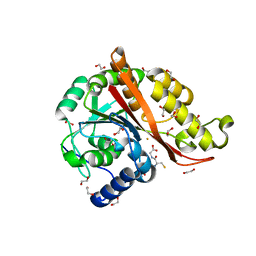 | |
7L5C
 
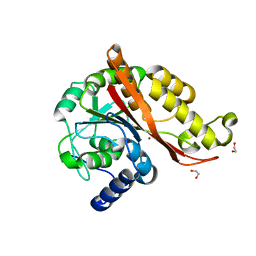 | |
6UXE
 
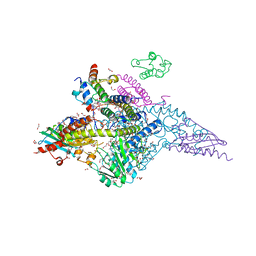 | | Structure of the human mitochondrial desulfurase complex Nfs1-ISCU2(M140I)-ISD11 with E.coli ACP1 at 1.57 A resolution showing flexibility of N terminal end of ISCU2 | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ... | | Authors: | Boniecki, M.T, Cygler, M. | | Deposit date: | 2019-11-07 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
To Be Published
|
|
6W1D
 
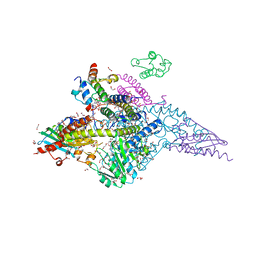 | | Structure of human mitochondrial complex Nfs1-ISCU2 (WT)-ISD11 with E.coli ACP1 at 1.8 A resolution (NIAU)2 | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ... | | Authors: | Boniecki, M.T, Cygler, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
To Be Published
|
|
6WI2
 
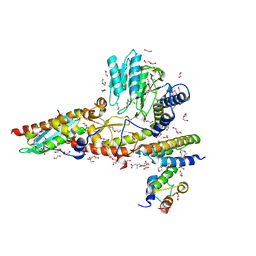 | |
6WIH
 
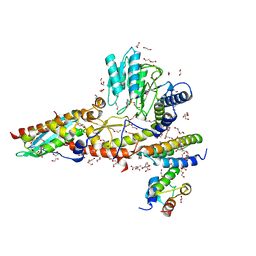 | | N-terminal mutation of ISCU2 (L35H36) traps Nfs1 Cys loop in the active site of ISCU2 without metal present. Structure of human mitochondrial complex Nfs1-ISCU2(L35H36)-ISD11 with E.coli ACP1 at 1.9 A resolution (NIAU)2. | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Acyl carrier protein, ... | | Authors: | Boniecki, M.T, Cygler, M. | | Deposit date: | 2020-04-09 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
To Be Published
|
|
3ZIU
 
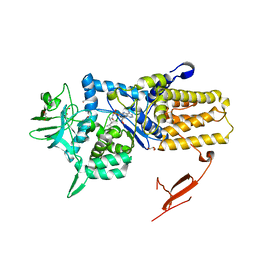 | | Crystal structure of Mycoplasma mobile Leucyl-tRNA Synthetase with Leu-AMS in the active site | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, LEUCYL-TRNA SYNTHETASE | | Authors: | Li, L, Palencia, A, Lukk, T, Li, Z, Luthey-Schulten, Z.A, Cusack, S, Martinis, S.A, Boniecki, M.T. | | Deposit date: | 2013-01-10 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leucyl-tRNA Synthetase Editing Domain Functions as a Molecular Rheostat to Control Codon Ambiguity in Mycoplasma Pathogens.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6MCQ
 
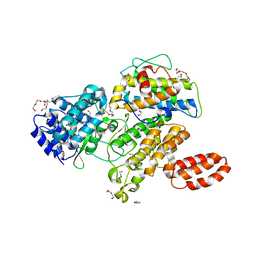 | | L. pneumophila effector kinase LegK7 in complex with human MOB1A | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, LegK7, ... | | Authors: | Beyrakhova, K.A, Xu, C, Boniecki, M.T, Cygler, M. | | Deposit date: | 2018-09-01 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TheLegionellakinase LegK7 exploits the Hippo pathway scaffold protein MOB1A for allostery and substrate phosphorylation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6MCP
 
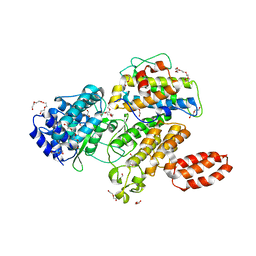 | | L. pneumophila effector kinase LegK7 (AMP-PNP bound) in complex with human MOB1A | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, LegK7, ... | | Authors: | Beyrakhova, K.A, Xu, C, Boniecki, M.T, Cygler, M. | | Deposit date: | 2018-09-01 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | TheLegionellakinase LegK7 exploits the Hippo pathway scaffold protein MOB1A for allostery and substrate phosphorylation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5UAM
 
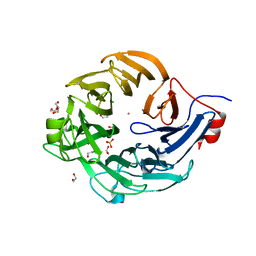 | | Structure of a new family of Polysaccharide lyase PL25-Ulvanlyase. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ulaganathan, T.S, Boniecki, M.T, Cygler, M. | | Deposit date: | 2016-12-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Ulvan-Degrading Polysaccharide Lyase Family: Structure and Catalytic Mechanism Suggests Convergent Evolution of Active Site Architecture.
ACS Chem. Biol., 12, 2017
|
|
5UFK
 
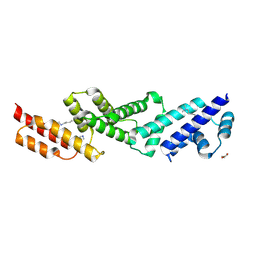 | | Structure of the effector protein SidK (lpg0968) from Legionella pneumophila | | Descriptor: | GLYCEROL, effector protein SidK | | Authors: | Beyrakhova, K, Xu, C, Boniecki, M.T, Cygler, M. | | Deposit date: | 2017-01-04 | | Release date: | 2017-05-10 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
5UF5
 
 | | Structure of the effector protein SidK (lpg0968) from Legionella pneumophila (domain-swapped dimer) | | Descriptor: | GLYCEROL, effector protein SidK | | Authors: | Beyrakhova, K, Xu, C, Boniecki, M.T, Cygler, M. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
5JG4
 
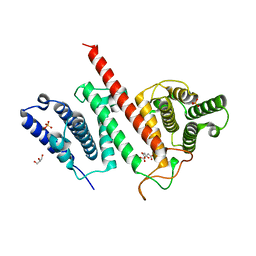 | | Structure of the effector protein LpiR1 (Lpg0634) from Legionella pneumophila | | Descriptor: | CITRATE ANION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Beyrakhova, K, van Straaten, K, Cygler, M. | | Deposit date: | 2016-04-19 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Investigations of the Effector Protein LpiR1 from Legionella pneumophila.
J.Biol.Chem., 291, 2016
|
|
5FIA
 
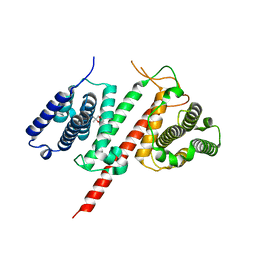 | | Structure of the effector protein LpiR1 (Lpg0634) from Legionella pneumophila | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LpiR1 | | Authors: | Beyrakhova, K, van Straaten, K, Cygler, M. | | Deposit date: | 2015-12-22 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Investigations of the Effector Protein LpiR1 from Legionella pneumophila.
J.Biol.Chem., 291, 2016
|
|
6DEH
 
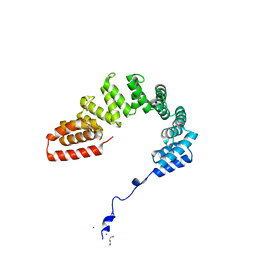 | | Structure of LpnE Effector Protein from Legionella pneumophila (sp. Philadelphia) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Voth, K, Chung, I.Y.W, van Straaten, K.E, Cygler, M. | | Deposit date: | 2018-05-11 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Legionella effector protein LpnE provides insights into its interaction with Oculocerebrorenal syndrome of Lowe (OCRL) protein.
FEBS J., 286, 2019
|
|
5UAS
 
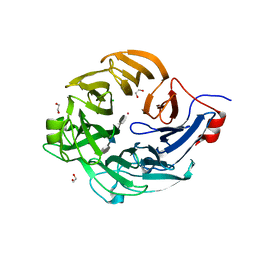 | | Structure of a new family of Polysaccharide lyase PL25-Ulvanlyase bound to -[GlcA(1-4)Rha3S]- | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose, CHLORIDE ION, ... | | Authors: | Ulaganathan, T.S, Cygler, M. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Ulvan-Degrading Polysaccharide Lyase Family: Structure and Catalytic Mechanism Suggests Convergent Evolution of Active Site Architecture.
ACS Chem. Biol., 12, 2017
|
|
5VOX
 
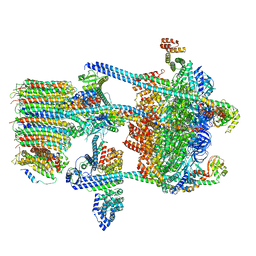 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 1) | | Descriptor: | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
5VOZ
 
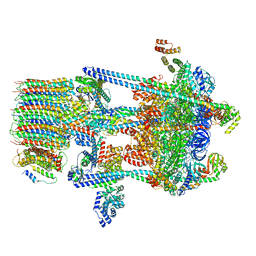 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 3) | | Descriptor: | Uncharacterized protein, V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
5VOY
 
 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 2) | | Descriptor: | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
